A List of Heredity Related Molecules: 2
This is the contiuned text of the previous one.
- Transcriptional repressors: Proteins that bind to DNA and inhibit gene expression by preventing the transcriptional machinery from accessing the DNA or recruiting chromatin-modifying complexes.
- Transcriptional enhancers: DNA sequences that can enhance gene expression by binding specific transcription factors and promoting the assembly of the transcriptional machinery.
- DNA-binding co-factors: Proteins that interact with transcription factors and assist in their binding to specific DNA sequences, modulating gene expression.
- DNA architectural proteins: Proteins that help organize the three-dimensional structure of DNA, bringing distant DNA elements into spatial proximity and influencing gene regulation.
- RNA export factors: Proteins that facilitate the export of RNA molecules from the nucleus to the cytoplasm, allowing them to engage in translation and other cellular processes.
- RNA decay enzymes: Enzymes that degrade RNA molecules, controlling their abundance and eliminating unwanted or defective RNA.
- RNA interference (RNAi) effectors: Proteins involved in the RNAi pathway, which uses small RNA molecules to silence gene expression by targeting and degrading complementary mRNA.
- Transcription termination factors: Proteins that participate in the termination of transcription, helping to release RNA polymerase and the nascent RNA from the DNA template.
- Chromosome remodeling enzymes: Enzymes that modify the structure and composition of chromosomes, contributing to chromatin organization, gene regulation, and DNA repair.
- DNA sliding clamps: Proteins that encircle DNA strands and provide a platform for other proteins, ensuring processive DNA synthesis during replication or repair
- RNA-binding proteins: Proteins that interact with RNA molecules, influencing their stability, localization, processing, and translation.
- RNA polymerase II: An enzyme responsible for transcribing protein-coding genes into pre-mRNA, a precursor molecule for mRNA.
- DNA recombination factors: Proteins involved in the rearrangement of DNA sequences, such as RecA, which catalyzes homologous recombination.
- Chromatin immunoprecipitation (ChIP) factors: Antibodies or proteins used in the ChIP technique to identify DNA regions bound by specific proteins, providing insights into gene regulation.
- Transcriptional activators: Proteins that bind to DNA and enhance gene expression by recruiting co-activators and facilitating the assembly of the transcriptional machinery.
- RNA-binding protein complexes: Multi-subunit complexes that interact with RNA molecules, influencing various aspects of RNA metabolism and function.
- DNA glycosylases: Enzymes that remove damaged or incorrect nucleotides from DNA through base excision repair, maintaining DNA integrity.
- RNA helicase complexes: Multi-protein complexes that unwind RNA molecules, enabling their processing, translation, and ribosome assembly.
- DNA replication terminators: Sequences that signal the termination of DNA replication, ensuring the accurate duplication of the genome.
- RNA processing enzymes: Enzymes involved in the modification and maturation of RNA molecules, such as splicing, capping, and polyadenylation.
- Nucleosome remodeling enzymes: Enzymes that reposition, evict, or alter the structure of nucleosomes, modulating chromatin accessibility and gene expression.
- DNA-binding architectural proteins: Proteins that shape and organize DNA within the nucleus, contributing to higher-order chromatin structure and gene regulation.
- RNA surveillance complexes: Protein complexes that monitor and degrade aberrant RNA molecules, preventing their translation or potential harm to the cell.
- DNA damage response effectors: Proteins that execute the cellular response to DNA damage, activating repair pathways, cell cycle checkpoints, or apoptosis if damage is severe.
- Histone acetyltransferase complexes: Multi-protein complexes that add acetyl groups to histone proteins, modulating chromatin structure and gene expression.
- RNA editing factors: Proteins or enzymes that catalyze post-transcriptional modifications of RNA, altering the RNA sequence or structure.
- Transcriptional co-repressors: Proteins that interact with transcription factors to inhibit gene expression by recruiting chromatin-modifying complexes or blocking the assembly of the transcriptional machinery.
- DNA replication restart factors: Proteins involved in restarting stalled DNA replication forks, ensuring the completion of DNA replication.
- DNA translocases: Proteins that move along DNA strands, participating in processes such as DNA repair, recombination, or chromatin remodeling.
- RNA transport complexes: Protein complexes responsible for transporting RNA molecules from the nucleus to specific subcellular locations for localized translation or storage.
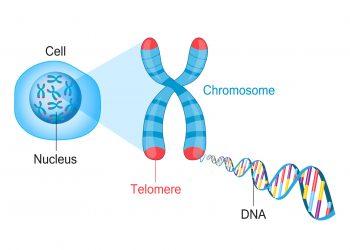
- Telomere-associated complexes: Protein complexes that interact with telomeres, contributing to their structure, stability, and regulation of telomere length.
- RNA editing regulators: Proteins that modulate RNA editing processes, controlling the extent or specificity of RNA modifications.
- DNA base excision repair factors: Proteins involved in the repair of damaged or modified DNA bases, maintaining genome integrity.
- Transcriptional elongation factors: Proteins that promote RNA polymerase progression during transcription, facilitating efficient transcriptional elongation.
- RNA chaperone complexes: Multi-protein complexes that assist in the folding, stability, and function of RNA molecules.
- Chromatin insulator proteins: Proteins that establish boundaries within the genome, preventing the spread of chromatin modifications and regulating gene expression.
- DNA damage bypass factors: Proteins that enable replication to proceed past DNA lesions or obstacles, ensuring the completion of DNA replication.
- RNA export adaptors: Proteins that mediate the interaction between RNA molecules and the export machinery, facilitating their transport out of the nucleus.
- DNA supercoiling regulators: Proteins that modulate the degree of DNA supercoiling, influencing DNA structure and accessibility.
- Transcriptional co-activator complexes: Multi-protein complexes that enhance gene expression by facilitating the assembly of the transcriptional machinery and promoting chromatin remodeling.
- DNA gyrase: An enzyme that introduces negative supercoils into DNA, playing a role in DNA topology and regulation of gene expression.
- RNA polymerase III: An enzyme responsible for transcribing transfer RNA (tRNA) genes and some small non-coding RNA genes.
- DNA replication primase: An enzyme that synthesizes short RNA primers during DNA replication to initiate DNA synthesis.
- DNA mismatch recognition proteins: Proteins that identify and bind to mismatched base pairs during DNA repair processes.
- RNA-induced transcriptional silencing (RITS) complex: A protein complex involved in RNA-mediated gene silencing by guiding histone modifications to target loci.
- RNA exonucleases: Enzymes that degrade RNA molecules by removing nucleotides from the ends.
- DNA replication processivity factors: Proteins that increase the processivity of DNA polymerases during DNA replication by tethering them to the DNA template.
- RNA 3′ end processing factors: Proteins that facilitate the cleavage and polyadenylation of the 3′ ends of RNA molecules during RNA processing.
- DNA resection enzymes: Enzymes that trim and process DNA ends during DNA repair processes, preparing them for recombination or repair synthesis.
- RNA export receptors: Proteins that mediate the transport of RNA molecules across the nuclear envelope for subsequent cytoplasmic functions.
- DNA polymerase switching factors: Proteins that facilitate the switching of DNA polymerases during DNA replication to accommodate different replication contexts.
- RNA editing substrates: RNA molecules that undergo post-transcriptional editing, resulting in changes to the RNA sequence or structure.
- DNA unwinding proteins: Proteins that assist in the unwinding of DNA helices during DNA replication, transcription, or repair.
- RNA editing cofactors: Proteins or RNA molecules that interact with RNA editing enzymes to enhance or regulate the editing process.
- DNA double-strand break repair factors: Proteins involved in repairing DNA double-strand breaks through mechanisms such as homologous recombination or non-homologous end joining.
- RNA interference (RNAi) suppressors: Proteins produced by viruses that inhibit the RNAi pathway, allowing the virus to evade host defense mechanisms.
- DNA replication checkpoint proteins: Proteins that monitor DNA replication progress and coordinate cell cycle progression to ensure accurate DNA synthesis.
- RNA editing regulators: Proteins that modulate the extent or specificity of RNA editing, influencing the type and frequency of edited sites.
- DNA polymerase epsilon: An enzyme involved in DNA replication that synthesizes the leading strand and participates in DNA repair processes.
- RNA silencing amplification complex (RDRC): A complex involved in amplifying small RNA signals and promoting the production of secondary small interfering RNAs (siRNAs).
- DNA end protection factors: Proteins that prevent the degradation or inappropriate processing of DNA ends during DNA repair.
- RNA export adaptors: Proteins that facilitate the recognition and export of specific RNA molecules from the nucleus to the cytoplasm.
- DNA interstrand crosslink repair factors: Proteins involved in repairing interstrand crosslinks, which covalently link the two DNA strands and can impede DNA transactions.
- RNA cap methyltransferases: Enzymes that methylate the 5′ cap structure of mRNA, influencing mRNA stability, translation, and splicing.
- DNA replication origin recognition proteins: Proteins that recognize and bind to specific DNA sequences to initiate DNA replication at replication origins.
- RNAi amplification factors: Proteins that amplify the RNAi response by enhancing the production or stability of small interfering RNAs (siRNAs27. DNA damage response phosphatases: Enzymes that dephosphorylate proteins involved in the DNA damage response, regulating their activity and signaling.
- RNA decapping enzymes: Enzymes that remove the 5′ cap structure from mRNA molecules, leading to mRNA degradation or regulation of translation.
- DNA replication helicase loaders: Proteins that assist in the loading of DNA helicases onto DNA during DNA replication initiation.
- RNA splicing regulators: Proteins that modulate alternative splicing of pre-mRNA molecules, influencing protein isoform diversity and gene expression.
- DNA alkylation repair enzymes: Enzymes involved in repairing DNA damage caused by the addition of alkyl groups, maintaining genome integrity.
- RNA editing quality control factors: Proteins that ensure the accuracy and fidelity of RNA editing processes, preventing errors or aberrant modifications.
- DNA damage response ubiquitin ligases: Enzymes that add ubiquitin molecules to proteins involved in the DNA damage response, regulating their stability, localization, or activity.
- RNA granule assembly factors: Proteins that contribute to the assembly and dynamics of RNA granules, regulating RNA localization, translation, and storage.
- DNA topoisomerase inhibitors: Small molecules that inhibit the activity of DNA topoisomerases, interfering with DNA topology and replication.
- RNA export adaptors: Proteins that facilitate the recognition and export of specific RNA molecules from the nucleus to the cytoplasm.
- DNA double-strand break sensors: Proteins that recognize and signal the presence of DNA double-strand breaks, initiating DNA repair pathways.
- RNA methylation readers: Proteins that recognize and bind to methylated RNA, influencing RNA stability, translation, or processing.
- DNA replication checkpoint kinases: Protein kinases that monitor DNA replication and activate signaling pathways to halt the cell cycle in response to replication stress.
- RNA editing quality control factors: Proteins that ensure the accuracy and fidelity of RNA editing processes, preventing errors or aberrant modifications.
- DNA unwinding regulators: Proteins that modulate the activity or processivity of DNA helicases during DNA replication, repair, or recombination.
- RNA export adaptors: Proteins that facilitate the recognition and export of specific RNA molecules from the nucleus to the cytoplasm.
- DNA polymerase proofreading subunits: Subunits of DNA polymerases that possess exonuclease activity, proofreading newly synthesized DNA strands for errors.
- RNA decay factors: Proteins that facilitate the degradation of RNA molecules, regulating their turnover and controlling gene expression.
- DNA damage tolerance factors: Proteins that allow DNA replication to continue in the presence of DNA lesions, ensuring completion of DNA synthesis.
- RNA surveillance factors: Proteins that monitor and degrade aberrant RNA molecules, preventing the accumulation of faulty transcripts.
- DNA repair helicases: Helicase enzymes that unwind DNA structures during DNA repair processes, facilitating access to damaged sites.
- RNA exosome complex: A multi-subunit complex involved in RNA processing and degradation, ensuring RNA quality control and turnover.
- DNA damage response adaptors: Proteins that mediate protein-protein interactions and facilitate the assembly of signaling complexes in response to DNA damage.
- RNA helicase inhibitors: Small molecules that inhibit the activity of RNA helicases, modulating RNA processing, translation, or RNA-protein interactions.
- DNA replication fork protection factors: Proteins that stabilize and protect replication forks during DNA replication, preventing fork collapse and promoting efficient DNA synthesis.
- RNA silencing suppressors: Proteins produced by viruses to counteract the host RNA silencing defense mechanisms, promoting viral replication and spread.
- DNA repair endonucleases: Enzymes that cleave DNA at specific sites during DNA repair processes, facilitating the removal of damaged or incorrect DNA segments.
- RNA export adaptors: Proteins that mediate the recognition and export of specific RNA molecules from the nucleus to the cytoplasm.
- DNA replication clamp loaders: Proteins that load sliding clamps onto DNA during DNA replication, enhancing the processivity of DNA polymerases.
- RNA localization factors: Proteins that guide specific RNA molecules to distinct subcellular locations, influencing localized gene expression and cellular processes.
- DNA replication fork restart factors: Proteins that aid in the restart of stalled replication forks, ensuring the completion of DNA replication.
- RNA silencing effectors: Proteins involved in the RNA silencing pathway that recognize and target specific RNA molecules for degradation or translational repression.
- DNA repair end processing factors: Proteins that process and prepare DNA ends for repair, enabling proper rejoining and restoration of genomic integrity.
- RNA turnover factors: Proteins that regulate the degradation of RNA molecules, controlling their abundance and turnover rate.
- DNA helicase inhibitors: Small molecules that inhibit the activity of DNA helicases, disrupting DNA replication, repair, or recombination processes.
- RNA export adaptors: Proteins that facilitate the recognition and export of specific RNA molecules from the nucleus to the cytoplasm.
- DNA replication origin firing factors: Proteins that promote the initiation of DNA replication at specific origins, ensuring accurate and timely DNA synthesis.
- RNA surveillance complexes: Protein complexes involved in the surveillance and elimination of aberrant or defective RNA molecules.
- DNA repair exonucleases: Enzymes that degrade DNA from the ends, playing a role in DNA repair pathways, such as resection or proofreading.
- RNAi machinery components: Proteins involved in the RNA interference pathway, including Dicer, Argonaute, and other factors that mediate small RNA processing and silencing.
- DNA replication termination factors: Proteins that facilitate the termination of DNA replication, ensuring proper completion and disengagement of replication machinery.
- RNA localization signals: Specific RNA sequences or structures that guide RNA molecules to distinct subcellular locations for localized functions.
- DNA damage response phosphodiesterases: Enzymes that remove DNA adducts or damaged nucleotides during DNA repair processes, restoring the integrity of the DNA molecule.
- RNA-binding domain proteins: Proteins that contain specific RNA-binding domains, allowing them to interact with RNA molecules and modulate their processing or function.
- DNA repair-associated chromatin modifiers: Proteins that modify chromatin structure during DNA repair processes, facilitating access to damaged DNA and promoting repair efficiency.
- RNA degradation enzymes: Enzymes involved in the degradation of RNA molecules, participating in pathways such as nonsense-mediated decay or RNA turnover.
- DNA replication initiation factors: Proteins that initiate the assembly of the DNA replication machinery at replication origins, marking the start of DNA synthesis.
- RNA turnover machinery components: Proteins involved in the degradation and turnover of RNA molecules, including exonucleases, decapping enzymes, and exosome components.
- DNA translocation motors: Motor proteins that translocate along DNA strands, contributing to processes such as DNA repair, recombination, or replication.
- RNA-induced epigenetic modifiers: RNA molecules that can induce changes in DNA or chromatin structure, leading to heritable epigenetic modifications.
- DNA repair-associated ATPases: ATP-dependent enzymes that participate in DNA repair processes, providing energy for DNA strand separation, remodeling, or nucleotide excision.
- RNA processing endonucleases: Enzymes that cleave RNA molecules internally during RNA processing79. RNA interference enhancers: Proteins or molecules that enhance the efficiency or potency of the RNA interference pathway, increasing gene silencing effects.
- DNA replication termination factors: Proteins that facilitate the termination of DNA replication at specific sites, ensuring proper completion of DNA synthesis.
- RNA stability factors: Proteins that regulate the stability of RNA molecules, influencing their degradation rates and half-lives.
- DNA repair-associated chromatin remodelers: Proteins that remodel chromatin structure during DNA repair processes, allowing access to damaged DNA sites.
- RNA surveillance sensors: Proteins that detect aberrant RNA molecules, triggering RNA degradation or quality control mechanisms.
- DNA replication processivity factors: Proteins that enhance the processivity of DNA polymerases during DNA replication, preventing premature dissociation from the DNA template.
- RNA splicing enhancers: Proteins or RNA sequences that promote the recognition and inclusion of specific exons during RNA splicing.
- DNA replication initiation regulators: Proteins that control the timing and frequency of DNA replication initiation, coordinating DNA synthesis with the cell cycle.
- RNA silencing amplification factors: Proteins that amplify small RNA signals, leading to the production of secondary small interfering RNAs (siRNAs) and reinforcement of gene silencing.
- DNA repair-associated ubiquitin ligases: Enzymes that add ubiquitin molecules to proteins involved in DNA repair processes, regulating their stability, interactions, or subcellular localization.
- RNA editing substrates: RNA molecules that undergo post-transcriptional editing, resulting in changes to their sequence, structure, or functional properties.
- DNA replication licensing regulators: Proteins that control the establishment and maintenance of DNA replication licensing, ensuring accurate and controlled DNA synthesis.
- RNA transport adaptors: Proteins that facilitate the transport of specific RNA molecules to distinct cellular compartments or structures.
- DNA repair-associated ATPases: ATP-dependent enzymes that participate in DNA repair processes, facilitating DNA strand separation, remodeling, or resolution of DNA structures.
- RNA decay regulators: Proteins that modulate the decay rates or degradation pathways of specific RNA molecules, influencing their turnover and abundance.
- DNA replication checkpoint effectors: Proteins that transmit and execute signals in response to DNA replication stress, halting the cell cycle or activating repair mechanisms.
- RNA modification enzymes: Enzymes that add or remove chemical modifications, such as methyl or acetyl groups, to RNA molecules, influencing their stability, function, or interactions.
- DNA repair-associated kinases: Protein kinases that phosphorylate target proteins involved in DNA repair processes, regulating their activity, localization, or interactions.
- RNA processing modulators: Proteins that regulate RNA processing events, such as alternative splicing, polyadenylation, or RNA editing, influencing gene expression and RNA function.
- DNA replication licensing inhibitors: Small molecules or proteins that inhibit the formation or activity of DNA replication licensing complexes, controlling DNA replication initiation.
- RNA export adaptors: Proteins that recognize and bind to specific RNA molecules, facilitating their export from the nucleus to the cytoplasm.
- DNA repair-associated SUMO ligases: Enzymes that conjugate SUMO (small ubiquitin-like modifier) molecules to target proteins involved in DNA repair processes, modulating their function, interactions, or subcellular localization.
These additional molecules further expand the complexity and diversity of the molecular players involved in heredity, gene regulation, and cellular processes.
See also:
A List Of Heredity Related Molecules: What are the heredity related molecules ?
https://fineupme.com/what-are-heredity-related-molecules/